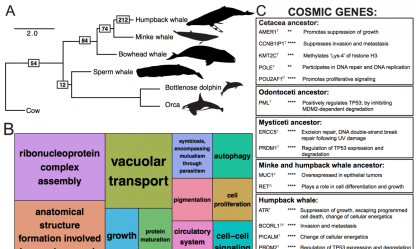
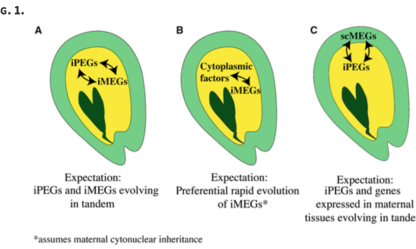
Inadvertent Paralog Inclusion Drives Artifactual Topologies and Timetree Estimates in Phylogenomics
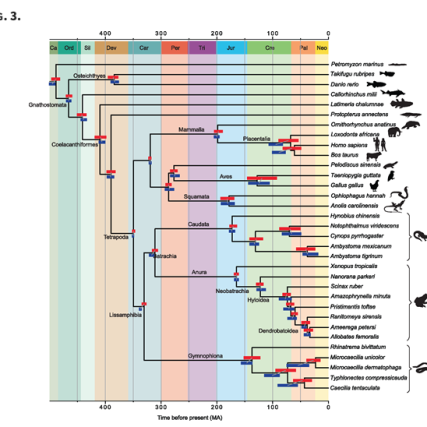
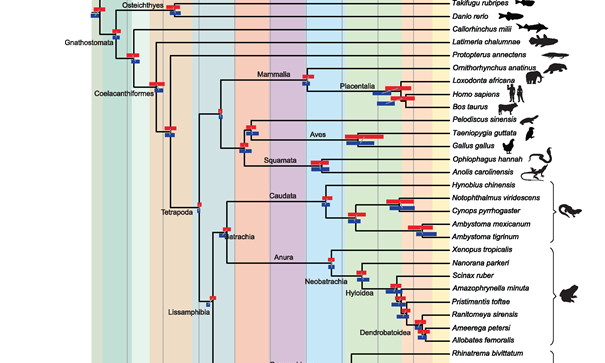
In work lead by Dr Karen Siu Ting she addresses an important question in phylogenomics using 18 amphibian species. She chose to work on this Lissamphibian group because there is significant conflict over their relationships to one another, and because transcriptomes are increasingly being used to overcome the lack of genomic data for this clade.…