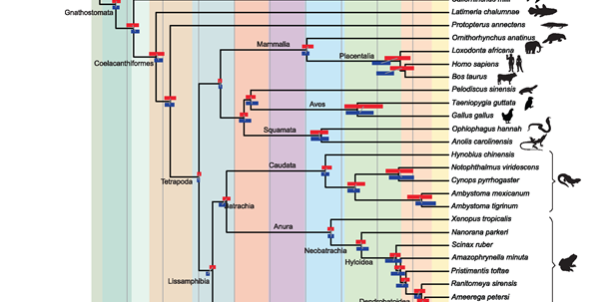
In work lead by Dr Karen Siu Ting she addresses an important question in phylogenomics using 18 amphibian species. She chose to work on this Lissamphibian group because there is significant conflict over their relationships to one another, and because transcriptomes are increasingly being used to overcome the lack of genomic data for this clade. In total 2656 putative single gene orthologs were identified and following the application of a novel paralog filtering approach phylogenetic reconstruction was carried out using Bayesian inference, maximum likelihood, and quartet-based supertrees. The paper describes how paralogs drive strongly supported conflicting hypotheses within the Lissamphibia (Batrachia and Procera) and older divergence time estimates even within groups where no variation in topology was observed. This is the first large-scale study to address the impact of orthology detection using transcriptomic data and emphasizes the importance of quality over quantity particularly for understanding relationships of poorly sampled taxa. Karen was a Marie Curie Postdoctoral Fellow between our group and Prof Creevey when she carried out this work and she is now based at Queen’s University in Belfast. Congratulations to Karen on a lovely piece of work.
Share This Article
Author: admin
This is the administrative account for the Computational & Molecular Evolutionary Biology Group at the University of Nottingham. Please browse through our site and contact us if you have any questions.